Statistical shape models (SSMs) are popular tools for model-based segmentation in 3D medical imaging. They are used to incorporate prior knowledge of organ shape variation in segmentation algorithms, and thereby greatly improve the quality of resulting segmentations.
(Matthias Kirschner)
Projects related to statistical shape models
- Construction of statistical shape models
- Visualization of statistical shape models
- Fully automatic organ segmentation
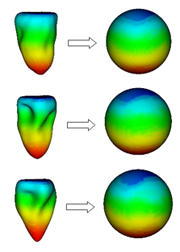
Construction of statistical shape models
SSMs are learned from a set of training shapes, which must be represented by a fixed number of landmarks point, and landmarks with the same index must describe the same anatomical feature. The problem of computing a landmark representation from a set of meshes with different number of points is known as the correspondence problem. Many state-of-the-art algorithms that solve this problem work on a parameter space representations of the training shapes.
The unit sphere is the natural parameter space for a great variety of organ shapes, as many organs have shapes with spherical topology, for example the liver, the kidney, the spleen or the ventricles of the heart.
In our research group, we have developed fast algorithms for spherical parameterization of sets of organ shapes. Our algorithms ensure that similar shape regions are mapped to the same regions on the unit sphere, such that the resulting parameterization are groupwise consistent. Moreover, area distortion is minimized, such that simple uniform sampling in the parameter space is sufficient to reconstruct all shapes with high quality. Our algorithms can be used to generate high quality SSMs directly, or to initialize state-of-the-art correspondence optimization algorithms close to an optimal solution.
M. Kirschner and S. Wesarg
Propagation of shape parameterisation for the construction of a statistical shape model of the left ventricle.
In: Proc. of Vision, Modeling, and Visualization Workshop (VMV), 2009.
M. Erdt, M. Kirschner and S. Wesarg
Simultaneous Segmentation and Correspondence Establishment for Statistical Shape Models.
In: Proc. of 2nd 3D Physiological Human Workshop 2009, Zermatt, Switzerland
M. Kirschner and S. Wesarg
Construction of Groupwise Consistent Shape Parameterizations by Propagation
In: Proc. of SPIE Medical Imaging 2010. 2010.
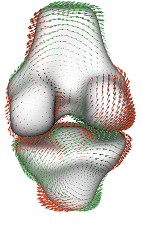
Visualization of statistical shape models
We have implemented a visualization tool for exploration of SSMs, which allows us to synthesize arbitrary shapes within the model's shape space. Our tool can visualize the eigenmodes of the model using glyphs, and facilitates the comparison of different shape models. The tool can be used to find model artifacts, and thus supports to “translate” quantitative evaluation results of point correspondence algorithms to qualitative results in form of images. This helps to get a better understanding of the benefits and limitations of different point correspondence algorithms.
M. Kirschner and S. Wesarg: Interactive Visualization of Statistical Shape Models. (Poster)
In: EG VCBM Workshop. 2010.
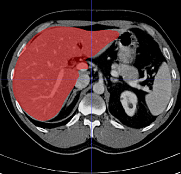
Fully automatic organ segmentation
We use statistical shape models for fully automatic segmentation. Our algorithms include organ detection, SSM initialization and state-of-the art Active Shape Model segmentation. Our work focuses on robust and accurate segmentation even in the case of large pathologies.
Marius Erdt, Matthias Kirschner, Sebastian Steger and Stefan Wesarg
Fast Automatic Liver Segmentation Combining Learned Shape Priors with Observed Shape Deviation.
IEEE International Symposium on Computer-Based Medical Systems, p. 249-254, 2010.
M. Kirschner and S. Wesarg
Active Shape Models unleashed. (to appear)
In: Proc. of SPIE Medical Imaging 2011.
M. Erdt, M. Kirschner, K. Drechsler, S. Wesarg, M. Hammon, A. Cavallaro
Automatic Pancreas Segmentation in Contrast Enhanced CT Data Using Learned Spatial Anatomy and Texture Descriptors (to appear)
In: Proc. ISBI 2011.


