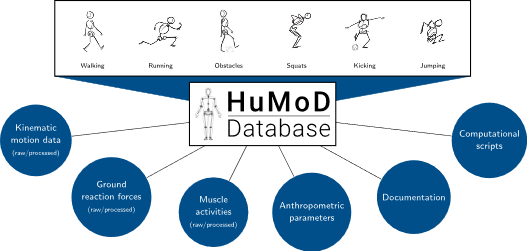
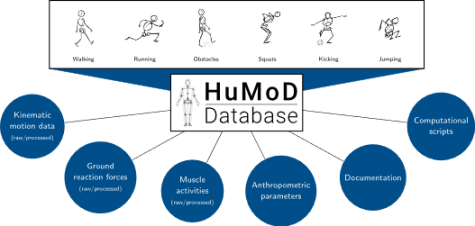
Documentation
The documentation (wird in neuem Tab geöffnet) provides detailed information about the structure and content of the data files as well as additional information about the subjects, motion protocol, measurement setup and data processing.
Examples
Example 1: Plot the right knee joint angle of the first female straight walking dataset
% Load female straight walking dataset
data = load(['A', filesep, '1.1.mat']);
% Find the index of the right knee joint in the cell array of trajectory
% labels where the individual letters in the label 'rKJZ_R' have the
% following meaning:
% r: revolute joint
% KJ: knee joint
% Z: rotation about local z-axis
% R: right side
indexKJ_R = find(strcmp(data.motion.trajectoryLabels, 'rKJZ_R'));
% Plot the right knee joint angle
timeKJ_R = 0:(1 / data.motion.frameRate):((data.motion.frames - 1) / data.motion.frameRate);
plot(timeKJ_R, data.motion.trajectory.q(indexKJ_R, :));
xlabel('Time in s');
ylabel('Right knee angle in rad');Example 2: Plot the left ankle joint acceleration, filtered activity of the left soleus muscle and left vertical ground reaction force of the 10th step in the first female straight running dataset
% Load female straight running dataset
data = load(['A', filesep, '2.1.mat']);
% Find the index of the left ankle joint in the cell array of trajectory
% labels where the individual letters in the label 'rAJZ_L' have the
% following meaning:
% r: revolute joint
% AJ: ankle joint
% Z: rotation about local z-axis
% L: left side
indexAJ_L = find(strcmp(data.motion.trajectoryLabels, 'rAJZ_L'));
% Find the index of the left soleus muscle in the cell array of muscle
% labels where the individual letters in the label 'SOL_L' have the
% following meaning:
% SOL: soleus muscle
% L: left side
indexSOL_L = find(strcmp(data.muscle.muscleLabels, 'SOL_L'));
% Compute start and end indices for the 10th step
stepStart = data.events.eventStart_L(10);
stepEnd = data.events.eventStart_L(11);
startIndexAJ_L = round(stepStart * data.motion.frameRate) + 1;
endIndexAJ_L = round(stepEnd * data.motion.frameRate) + 1;
startIndexSOL_L = round(stepStart * data.muscle.frameRate) + 1;
endIndexSOL_L = round(stepEnd * data.muscle.frameRate) + 1;
startIndexGRFY_L = round(stepStart * data.force.frameRate) + 1;
endIndexGRFY_L = round(stepEnd * data.force.frameRate) + 1;
% Plot the left ankle joint acceleration, filtered activity of the left
% soleus muscle and left vertical ground reaction force of the 10th step
subplot(1, 3, 1);
timeAJ_L = stepStart:(1 / data.motion.frameRate):stepEnd;
plot(timeAJ_L, data.motion.trajectory.ddqddt(indexAJ_L, startIndexAJ_L:endIndexAJ_L));
xlabel('Time in s');
ylabel('Left ankle acceleration in rad/s^2');
subplot(1, 3, 2);
timeSOL_L = stepStart:(1 / data.muscle.frameRate):stepEnd;
plot(timeSOL_L, 1e3 * data.muscle.activities.filtered(indexSOL_L, startIndexSOL_L:endIndexSOL_L));
xlabel('Time in s');
ylabel('Activity of the left soleus muscle in mV');
subplot(1, 3, 3);
timeGRFY_L = stepStart:(1 / data.force.frameRate):stepEnd;
plot(timeGRFY_L, data.force.grfY_L(startIndexGRFY_L:endIndexGRFY_L));
xlabel('Time in s');
ylabel('Left ground reaction force in N');License
The HuMoD Database is made available under the Open Database License v1.0. Any rights in individual contents of the database are licensed under the Database Contents License v1.0.
Scripts
The source code of the applied computational scripts is available in the HuMoD Database repository. The source code is licensed under the BSD 3-Clause License.
Citation
We provide the HuMoD Database in the hope that it might be useful to other researchers. Since a lot of time and effort has gone into creating the database, please cite the following publication (wird in neuem Tab geöffnet) if you are using processed or raw data or computational scripts provided in the context of the HuMoD Database in your own research.
inproceedings wojtusch2015humod,
author = Wojtusch, J and von Stryk, O,
title = HuMoD – A Versatile and Open Database for the Investigation, Modeling and Simulation of Human Motion Dynamics on Actuation Level,
booktitle = Proceedings of the IEEE-RAS International Conference on Humanoid Robots,
year = 2015,
address = Seoul,
doi = 10.1109/HUMANOIDS.2015.7363534,
pages = 74 -- 79,
publisher = IEEE
Statistics
The HuMoD Database website uses PIWIK for analysing anonymized visitor and download statatistics. This system respects the „Do Not Track“ preference and will not track your activities if you have specified „I do not want to be tracked“ in your web browser.
Contact
Please contact Janis Wojtusch for further information.
Changelog
| Version | Description |
| 1.3 |
Date: 2016.03.07 Commit: 3c29267 Updated release with two additional degrees of freedom in the torso segment and updated anthropometric parameters for the feet. |
| 1.2 |
Date: 2016.01.29 Commit: 077001a Updated release with updated anthropometric parameters and some general bug fixes. |
| 1.1 |
Date: 2015.09.29 Commit: ae46231 Updated release for IEEE-RAS International Conference on Humanoid Robots with updated anthropometric parameters and joint center estimates, additional degrees of freedom in the ankle joints and some general bug fixes. |
| 1.0 |
Date: 2015.05.04 Commit: c32ffbe Initial release of the HuMoD database and computational scripts. |


