We research visual analytis tools suporting biologic research, especially for evolutionary biology. We develop tools for comparison of phylogenetic trees, for analysis of mutation chains in proteins, for exploring mutual information in molecular evolution.
Cooperation: Prof. Kay Hamacher, TU Darmstadt, Computational Biology Group
Publications
- Visuall Comparison of Phylogenetic trees
- Visuall Analysis of Parameter influence on Phylogenetic trees
- Visual Analysis of Mutations in Proteins – Finding frequen biologic patterns in amino acid mutations
- Computing and Visually Analyzing Mutual Information in Molecular Co-Evolution
Within project work, we offer student jobs (HiWi), praktikas, bachelor and master theses, please contact Tatiana von Landesberger if interested.
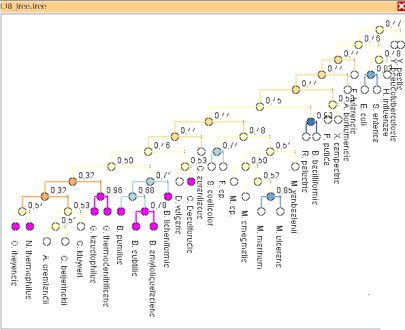
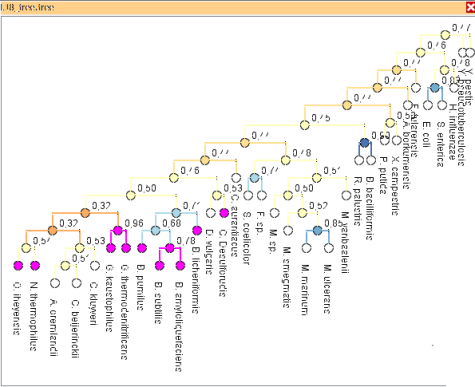
Visual Comparison of Phylogenetic Trees
In phylogeny analysis, multiple, differing trees need to be analyzed simultaneously in a comparative way -- in particular to highlight differences between them, which sometimes can be subtle.
We support comparison of several up to thousands of phylogenetic trees. We provide a set of interactive views comparing phylogenetic trees on various detail levels. The views incorporate information on similarity scores and allow highlighting of similar structures in multiple trees. They thereby reveal global and local tree similarities.
software, video, Paper (PDF)
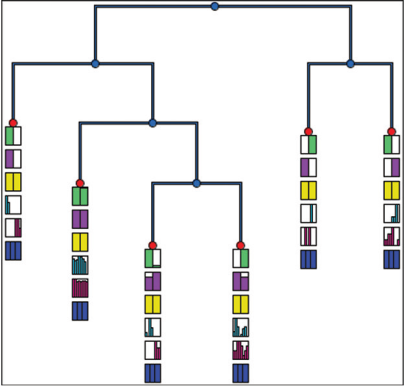
Visual Analysis of Parameter Influence on Phylogentic Trees
Evolutionary relationships between organisms are represented as phylogenetic trees inferred from multiple sequence alignments (MSAs).
The proposed approach clusters trees created with many MSA parameterizations. It displays the chosen parameters impact on the phylogenetic trees. This view offers interactive parameter exploration and automatic identi?cation of relevant parameters.
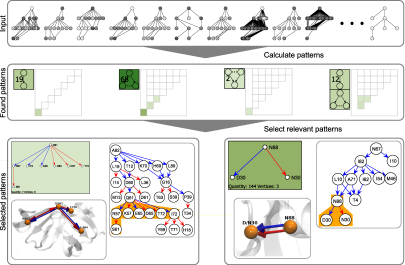
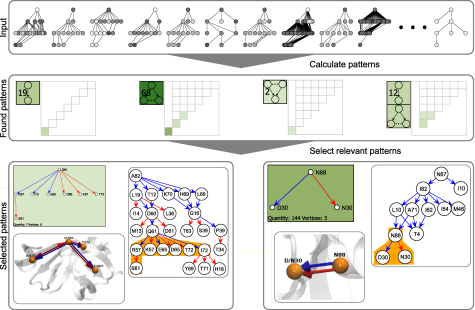
Visual Analysis of Patterns in Protein Mutations
Proteins are essential parts in all living organisms. They consist of sequences of amino acids. An interaction with reactive agent can stimulate a mutation at a specific position in the sequence.
We present a new system for the search and the exploration of frequent patterns (i.e., motifs) in mutation graphs. We present a fast pattern search algorithm specifically developed for finding biologically relevant patterns in many mutation graphs (i.e., many labeled acyclic directed graphs). Our visualization system allows an interactive exploration and comparison of the found patterns.
website, Paper (PDF)
Computing and Visually Analyzing Mutual Information in Molecular Co-Evolution
Selective pressure in molecular evolution leads to uneven distributions of amino acids and nucleotides. Infact one observes correlations among such constituents due to a large number of biophysical mechanisms To quantify these correlations the mutual information has proven most effective. The challenge is to navigate the large amount of data, which in a study for a typical protein cannot simply be plotted.
We developed a matrix visualization tool. It allows to take different views on the mutual information matrix by ltering, sorting, weighting, and zooming functionality. Thereby, the user can interactively navigate a huge matrix in real-time and search for e.g., patterns and unusual high or low values.
Visit the software page